Motivation and background on the Urfold, including synopses of what we’ve done so far (articles).
Motivation
The Urfold model/abstraction is motivated by our believe that it can help resolve some of the (seemingly mutually exclusive) dichotomies that have vexed the field of protein structural bioinformatics for many years– e.g., the nature of “fold space” (continuous vs discrete) as well as the issue of how (precisely, quantitatively) one might optimally (<= what does that mean, exactly?) perform as-objective-as-possible, statistically-principled comparative analyses across the universe of all protein 3D structures, the dichotomy in this case stemming from the algorithmic approaches to structure comparison—whether (a) one is cognizant of the topology/fold of the protein (i.e., the sequential order, along the polypeptide chain, in which SSEs [helices, strands] appear) or, alternatively, (b) adopt a more purely-geometric mindset, be agnostic of the topology, and instead worry only about the general ‘architecture’ of the 3D structure (which can be taken as the spatial arrangement of the ‘major’ SSEs). One issue is that many of these concepts have been rather subjectively formulated and heuristically implemented over the years (thresholds/cutoffs, etc. can alter even what one ‘defines’ as a protein ‘fold’; how do we ‘define’ a protein fold, in a statistically-principled way?). Largely for these reasons, we seek to utilize deep learning-based approaches to “define” the urfold (Eli’s “deepUrfold” project).
The following are synopses of our 3 papers thus far on the Urfold topic, in the order in which they’ve introduced the concept and then begun to develop it. We also have another paper currently in-prep, with a working title “Deep Generative Models of Protein Domain Structures Can Uncover Distant Relationships: Evidence for an ‘Urfold’?”.
1. “The Small β-Barrel Domain: A Survey-Based Structural Analysis” (https://pubmed.ncbi.nlm.nih.gov/30393050/)
This is a lengthy, tour-de-force style article (25+ pages & supplemental) from late 2018. It utilized both sledgehammers and scalpels to address such questions as “why/how do these small, β-rich protein structures [which seem to be functionally related, too] end up being classified [by hierarchical classification systems such as CATH, SCOP, etc.] differently– in some cases even quite differently? what gives?” Pursuit of those types of questions (i) occurred in a largely manual, old-fashioned manner, (ii) included the application of a heavy dose of domain-specific, human-expertise-based knowledge, and (iii) was limited in scope to proteins containing these so-called “small β-barrel” (SBB) domain. Indeed, the SBB is our ‘founder’ example of an urfold. Now, a major goal w/ our DeepUrfold project (and, indeed, NSF grant proposal) is to make the entire process (e.g., “process” = “identifying and discovering new urfolds”) more objectively-grounded (less heuristic), more precise, more rigorous (geometrically and statistically principled), and systematic (scalable/automatable, too, so we can deploy it across all known protein structures, allowing construction of a new map of the “protein structure universe”).
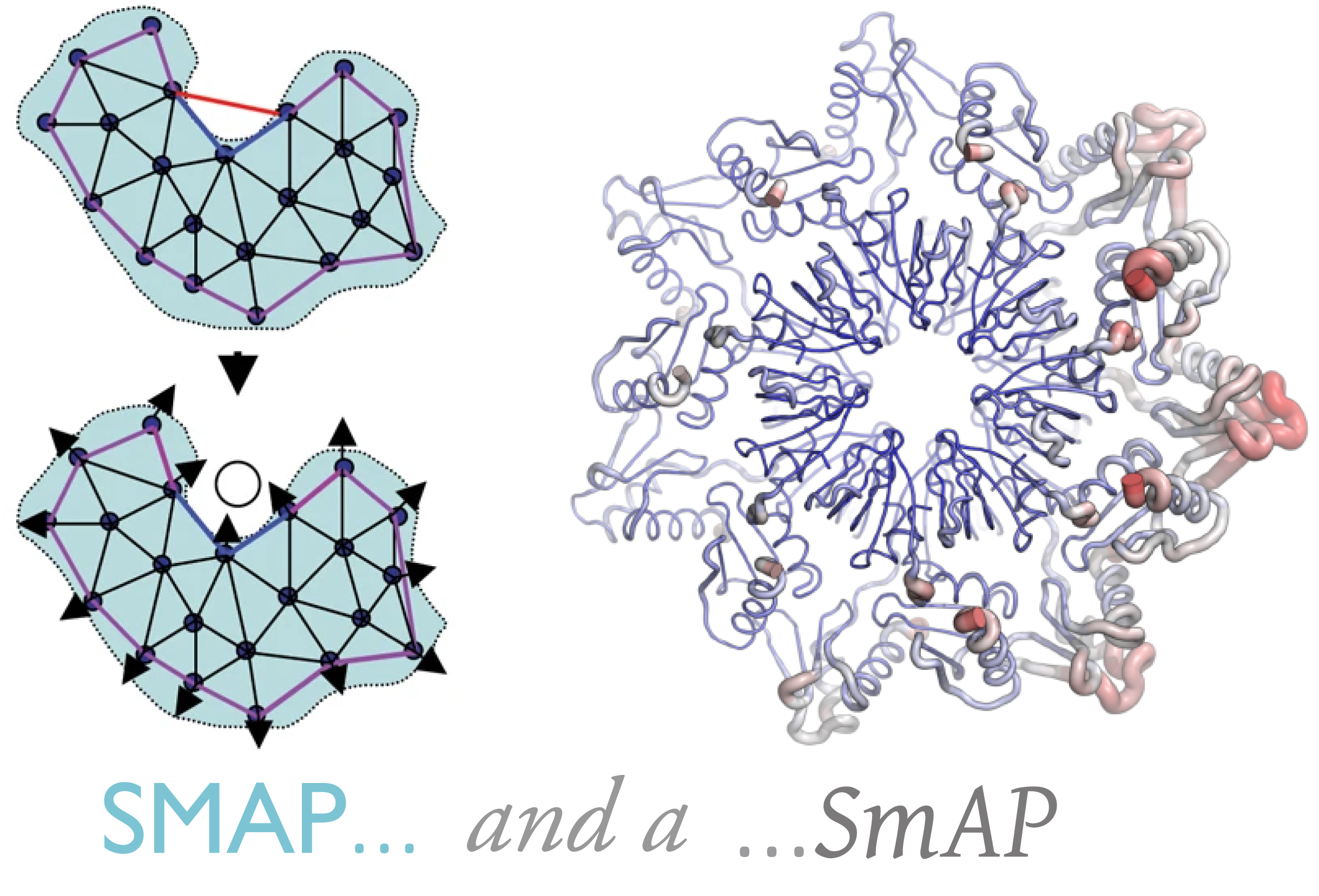
2. “The Urfold: Structural similarity just above the superfold level?” (https://onlinelibrary.wiley.com/doi/full/10.1002/pro.3742)
From late 2019, we offer a briefer and more tightly-focused work (vs the above SBB story), wherein we really begin to try and systematically define (or at least understand) what is an urfold? We provide what we believe are some further examples (beyond just the SBB), and we try to really delineate and articulate the concept that we believe to be really the key hallmark of the Urfold–namely, “architectural similarity despite topological variability”. (While that that may sound trite or not so profound outside the field of protein structural biology, it flies in the face of much conventional thinking about the patterns–ontologically speaking [see Figure 1]–of [dis]similarities amongst a collection of protein structures, and such a phenomenon wreaks havoc [and fundamentally undermines, I believe] the gold-standard protein structure classification schemes, like CATH and SCOP– those are predicated on hierarchical clustering [via different linkage algorithms, to make matters more confusing!]… and also they employ distance measures that are not true [mathematically proper] metrics… which, I believe, brings into question even the validity of a hierarchical clustering approach…). At any rate, the laundry list in the last sentence describes some of the reasons that have motivated our more recent adoption of Deep Learning as the way to approach this problem of protein structure analysis.
3. “Deep Learning of Protein Structural Classes: Any Evidence for an ‘Urfold’?” (https://arxiv.org/abs/2005.08443)
More recently, our preprint presents a hybrid deep learning architecture that consists of an autoencoder and 3D-CNNs for the structural (latent space) representations– using it, we find some tantalizing results that are at least consistent with what we had gleaned (manually, using human expertise) in the earlier work (such as the SBB, above). If you have time to look at any one of these articles, then I’d recommend starting with this one, in terms of our recent computational strategies. To really cut to the chase– in terms of what we’re interested in, re: picking your mind and considering potential downstream collaborations? – I’ll quote from our DeepUrfold paper (and highlight the most salient part): “………Here, we describe our training of Deep Learning models on protein domain structures (and their associated physicochemical properties) in order to evaluate classification properties at CATH’s “homologous superfamily” (SF) level. To achieve this, we have devised and applied an extension of image-classification methods and image segmentation techniques, utilizing a convolutional autoencoder model architecture. Our DL architecture allows models to learn structural features that, in a sense, ‘define’ different homologous SFs. We evaluate and quantify pairwise ‘distances’ between SFs by building one model per SF and comparing the loss functions of the models. Hierarchical clustering on these distance matrices provides a new view of protein interrelationships–a view that extends beyond simple structural/geometric similarity, and towards the realm of structure/function properties.”